3D multimodal muscle histology
Since Spring 2024
This was my first project in Dr. Thakor’s lab when I first joined as part of his lab’s work on vascularized denervated muscle targets.
The objective is to visualize and analyze structural properties of the muscle obtained through histology and correlate them to electrophysiological activity recorded in vivo.
The histology images were processed using different levels of image processing techniques, isolating the muscle and its shape from the rest. Key features of the tissue such as Neuromuscular Junctions (NMJs) or muscle fibers were identified either manually or using a region finding algorithm. The filtered images are then stacked based on the distance at which they were cut recreating the 3D model. Features are plotted on top of the model using the coordinates extracted from each histology layer. A convex hull is finally used to recreate the shape of the muscle.
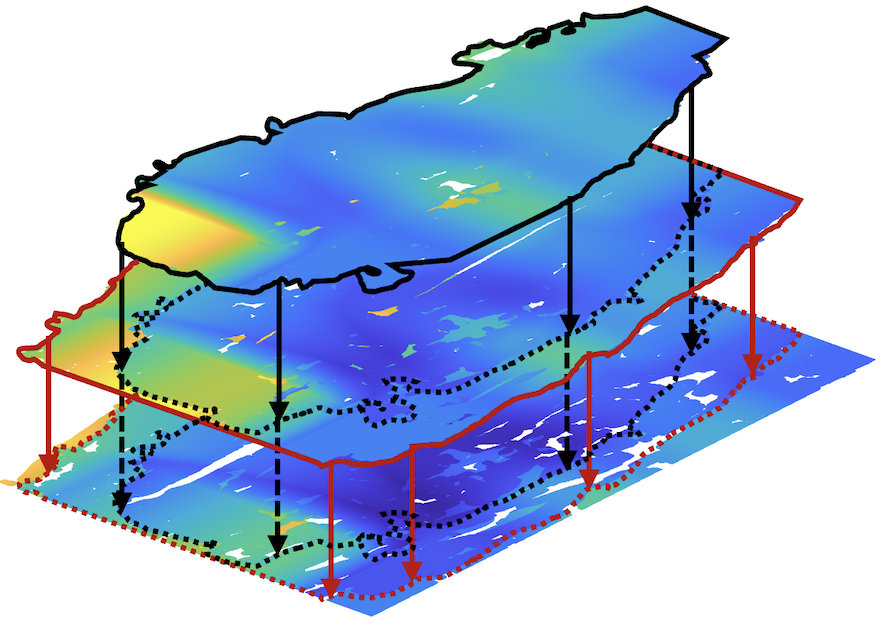
![]() Plotted are the NMJs of the muscle. The color indicates the level of maturation of the junctions.
Plotted are the NMJs of the muscle. The color indicates the level of maturation of the junctions.
![]()
Illustration of the analysis methods from feature extraction (J-K) to multi muscle distribution analysis using voxel method (L) and Density based method (M)
The objective is to visualize and analyze structural properties of the muscle obtained through histology and correlate them to electrophysiological activity recorded in vivo.
Creating 3D models
The histology images were processed using different levels of image processing techniques, isolating the muscle and its shape from the rest. Key features of the tissue such as Neuromuscular Junctions (NMJs) or muscle fibers were identified either manually or using a region finding algorithm. The filtered images are then stacked based on the distance at which they were cut recreating the 3D model. Features are plotted on top of the model using the coordinates extracted from each histology layer. A convex hull is finally used to recreate the shape of the muscle.
 Plotted are the NMJs of the muscle. The color indicates the level of maturation of the junctions.
Plotted are the NMJs of the muscle. The color indicates the level of maturation of the junctions.Finding trends
Once the features have been extracted these can be processed and analyzed in multiple ways, using voxel regions or density based analysis to identify trends across multiple muscles. In our case we used this to find reinnervation patterns in different types of muscle surgery, comparing a vascularized denervated muscle target and an intact muscle.
Illustration of the analysis methods from feature extraction (J-K) to multi muscle distribution analysis using voxel method (L) and Density based method (M)

Reconstruction and averaging of an NMJs in an intact muscle vs a reinnervated vascularized muscle target.
Overlapping EMG activity
Using high-density recording arrays, it is possible to measure the individual activation of a muscle’s motor units. By analyzing the resulting heatmaps and extracted features, the correlation between specific NMJs and motor units can be identified, providing insights into the relationship between structural and electrophysiological organization. This holds significant research value for understanding muscle diseases and their progression, as well as being able to understand better reinnervation patterns in muscles. This could be helpful also for prosthetic control in cases such as RPNI
Team
This work was greatly supported and completed thanks to the help of Manan Bhatt, Kiara Quinn, Siyu Wang and Pierce Perkins.